Code
3.12.2 (main, Feb 6 2024, 20:19:44) [Clang 15.0.0 (clang-1500.1.0.2.5)]
pompon version = 0.0.9
macOS-14.4.1-arm64-arm-64bitIn this case, we consider PES written by \[ V(x,y) = 4x^2 + 3y^2 + 2xy \]
3.12.2 (main, Feb 6 2024, 20:19:44) [Clang 15.0.0 (clang-1500.1.0.2.5)]
pompon version = 0.0.9
macOS-14.4.1-arm64-arm-64bitSampling from Boltzman distribution \(\exp(-\beta V)\) is anallytically achievable by using multivariate normal distribution.
def sample_ellipse(
N: int = 100, β: float = 1.0e01
) -> tuple[np.ndarray, np.ndarray]:
# sample from a 2D Gaussian
np.random.seed(0)
A = np.array([[4, 1], [1, 3]])
# Σ^-1 / 2 = A * β
Σ = np.linalg.inv(A * β * 2.0)
μ = np.array([0, 0])
x = np.random.multivariate_normal(μ, Σ, N)
return x[:, 0], x[:, 1]x = np.linspace(-1, 1, 100)
y = np.linspace(-1, 1, 100)
X, Y = np.meshgrid(x, y)
Z = ellipse_pes(X.flatten(), Y.flatten()).reshape(X.shape)
plt.contourf(X, Y, Z, 20, cmap="RdGy", vmax=9, vmin=0)
plt.colorbar()
x, y = sample_ellipse(N=100, β=1.0e01)
plt.scatter(x, y, label="β=1.e+01")
x, y = sample_ellipse(N=100, β=1.0e00)
plt.scatter(x, y, label="β=1.e+00")
plt.legend()
plt.show()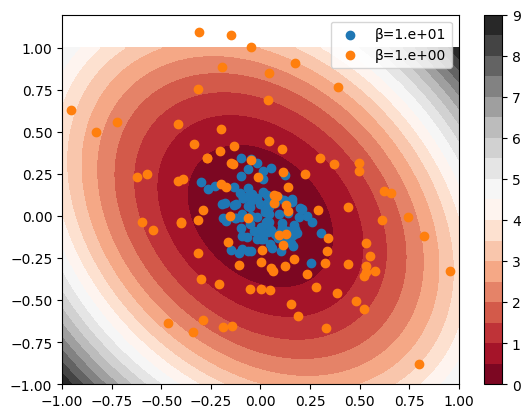
One constructs following NNMPO
You can change activation. See pompon.layers.activations for available activations.
Sample 256 points with temperature β=1.0
(179, 2)
(179, 1)
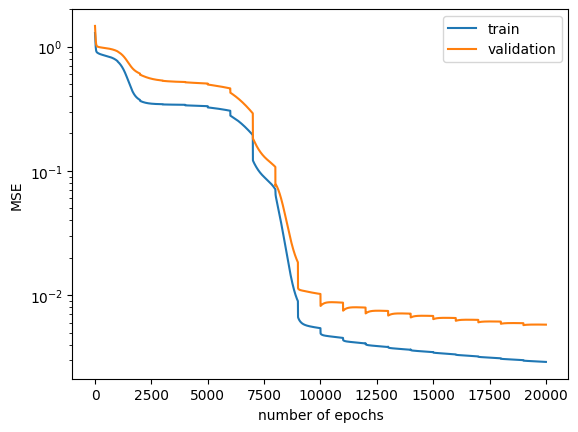
To grasp the optimization routine, one employs manual gradient descent (GD) rather than a trainer.
for i_epoch in tqdm(range(20_000)):
evaluate_loss(nnmpo, x_train, y_train, train_losses)
evaluate_loss(nnmpo, x_validate, y_validate, validation_losses)
params = nnmpo.grad(
x_train, y_train, basis_grad=True, coordinator_grad=True
)
for param in params:
param.data -= lr * param.grad
if (i_epoch + 1) % 1_000 == 0:
nnmpo.update_blocks_batch(x_train)
for _ in tqdm(range(5_000)):
params = nnmpo.grad(
x_train, y_train, twodot_grad=True, to_right=True
)
B: TwodotCore = params[0]
B.data -= lr_tt * B.grad
nnmpo.tt.norm.data *= (norm := jnp.linalg.norm(B.data))
B.data /= norm
W0, W1 = B.svd(truncation=0.99, rank=2)
nnmpo.tt.W0.data = W0.data
nnmpo.tt.W1.data = W1.data
norms.append(nnmpo.tt.norm.data)
tt_rank_epochs.append(nnmpo.tt.ranks)Explanation of the code:
Lines 1~3:
for i_epoch in tqdm(range(20_000)):
evaluate_loss(nnmpo, x_train, y_train, train_losses)
evaluate_loss(nnmpo, x_validate, y_validate, validation_losses)Performs training for 20,000 epochs. Calculates Mean Squared Error (MSE) every epoch and logs it.
Lines 4~6:
params = nnmpo.grad(x_train, y_train, basis_grad=True, coordinator_grad=True)
for param in params:
param.data -= lr * param.gradRetrieves gradients for parameters \(w\), \(b\), and \(U\) using automatic differentiation and optimizes them using gradient descent. The \(U\) is automatically retracted to the Stiefel manifold by calling - operator.
Lines 7~8:
Optimizes Tensor Train (TT) every 1000 epochs. Computes all TT basis for each batch.
Lines 9~12:
for _ in tqdm(range(5_000)):
params = nnmpo.grad(x_train, y_train, twodot_grad=True)
B = params[0] #
B.data -= lr_tt * B.gradTwodot core \(B\) is also optimized by using gradient descent. Here, gradients are calculated analytically.
Lines 13~17:
nnmpo.tt.norm.data *= (norm := jnp.linalg.norm(B.data))
B.data /= norm
W0, W1 = B.svd(truncation=0.99, rank=2)
nnmpo.tt.W0.data = W0.data
nnmpo.tt.W1.data = W1.dataAfter gradient descent optimization of Twodot core \(B\), Twodot core \(B\) is normalized and decomposed into two Onedot cores \(W\) using Singular Value Decomposition (SVD). It adaptively truncates small singular values and corresponding singular vectors, keeping only the top 99% contribution.
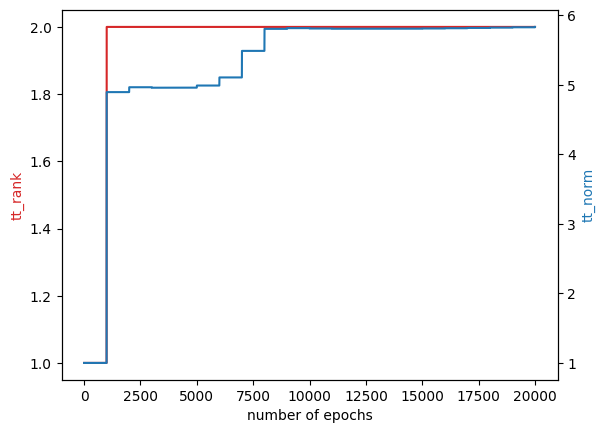
Why did we set rank=2?
The elliptic function has a 3-rank structure. \[ 4x^2 + 3y^2 + 2xy = \begin{bmatrix} 4x^2 & 2x & 1 \end{bmatrix} \begin{bmatrix} 1 \\ y \\ 3y^2 \end{bmatrix} \] However, if coordinator learns low-rank coordinates, \[ [q_1, q_2] = [x, y] U \] where \(U\) diagonalizes the Hessian matrix, \[ A = \begin{bmatrix} 8 & 1 \\ 1 & 6 \end{bmatrix} \] then, the elliptic function becomes \[ 4x^2 + 3y^2 + 2xy = \lambda_1 q_1^2 + \lambda_2 q_2^2 \] where \(\lambda_1\) and \(\lambda_2\) are eigenvalues of \(A\). Thus, we can rewrite the elliptic function as a 2-rank structure. \[ \begin{bmatrix} \lambda_1 & \lambda_2 q_1^2 \\ \end{bmatrix} \begin{bmatrix} q_2^2 \\ 1 \end{bmatrix} \]
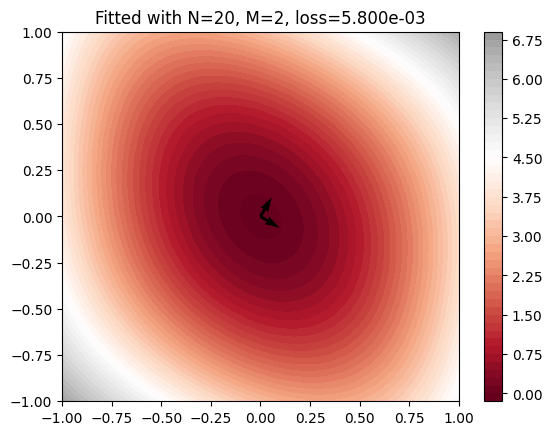
Visualize results
x = np.linspace(-1.0, 1.0, 100)
y = np.linspace(-1.0, 1.0, 100)
X, Y = np.meshgrid(x, y)
Z = nnmpo.forward(jnp.stack([X.flatten(), Y.flatten()], axis=1))[:, 0].reshape(
X.shape
)
plt.title(
f"Fitted with N={N}, "
+ f"M={max(tt_rank_epochs[-1])}, "
+ f"loss={validation_losses[-1]:.3e}"
)
plt.contourf(X, Y, Z, 50, cmap="RdGy", vmax=9, vmin=0)
plt.colorbar()
u = nnmpo.coordinator.U.data[0, :]
v = nnmpo.coordinator.U.data[1, :]
plt.quiver(0, 0, u[0], u[1])
plt.quiver(0, 0, v[0], v[1])
plt.show()