3.12.2 (main, Feb 6 2024, 20:19:44) [Clang 15.0.0 (clang-1500.1.0.2.5)]
pompon version = 0.0.9
macOS-14.4.1-arm64-arm-64bitH2CO molecute - optimize
This is a light example of how to optimize the molecular potential. For more sophistecated optimization, refer docs/notebook/_h2co_opt.py.
Environment
Import modules
Load data
Code
x_train = np.load("data/x_train.npy")
x_test = np.load("data/x_test.npy")
x_valid = np.load("data/x_validation.npy")
y_train = np.load("data/y_train.npy")
y_test = np.load("data/y_test.npy")
y_valid = np.load("data/y_validation.npy")
f_train = np.load("data/f_train.npy")
f_test = np.load("data/f_test.npy")
f_valid = np.load("data/f_validation.npy")
y_shift = -3112.1604302407663 # eV (mean)
y_min = -3113.4044750979383 # eV (eq. position)
plt.title("Train energy")
plt.violinplot(y_train + y_shift)
plt.axhline(y_min)
plt.ylabel("Energy [eV]")
plt.plot()
plt.show()
plt.title("Train coordinate")
plt.plot(np.arange(6), x_train.T)
plt.show()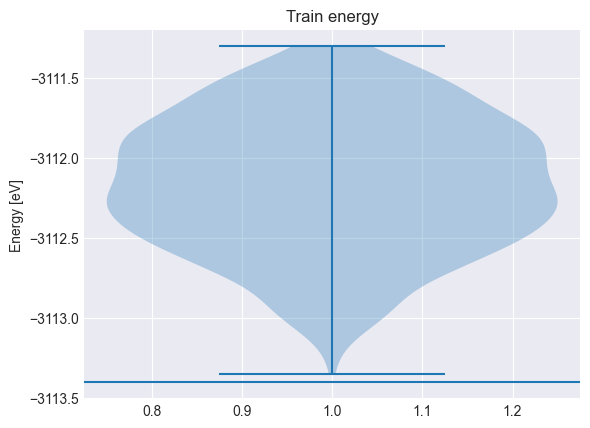
Consturct a model
MPO is encoded from sum of products functions. We start from SOP training, and then convert it to MPO.
Init signature: pompon.OneBody( input_size: int, hidden_size: int, basis_size: int, output_size: int = 1, w_scale: float = 1.0, b_scale: float = 1.0, w_dist: str = 'uniform', b_dist: str = 'linspace', x0: jax.Array | None = None, activation: str = 'moderate+silu', key: jax.Array | None = None, X_out: numpy.ndarray | None = None, fix_bias: bool = False, ) Docstring: Function given by sum of one-body functions $$ f(q_1, q_2, \ldots, q_f) = \sum_{p=1}^{f} \sum_{\rho_p} W_{\rho_p}^{(p)} \phi_{\rho_p}(w_{\rho_p}^{(p)} q_p+b_{\rho_p}^{(p)}) $$ File: ~/GitHub/Pompon/pompon/sop.py Type: ABCMeta Subclasses:
One-body regression
\[ f(q_1, q_2, \ldots, q_f) = \sum_{p=1}^{f} \sum_{\rho_p} W_{\rho_p}^{(p)} \phi_{\rho_p}(w_{\rho_p}^{(p)} (q_p-q^{\mathrm{ref}}_{\rho_p})+b_{\rho_p}^{(p)}) \] \[ \mathbf{q}-\mathbf{q}^{\mathbf{ref}} = (\mathbf{x} - \mathbf{x}^{\mathrm{ref}}) U \]
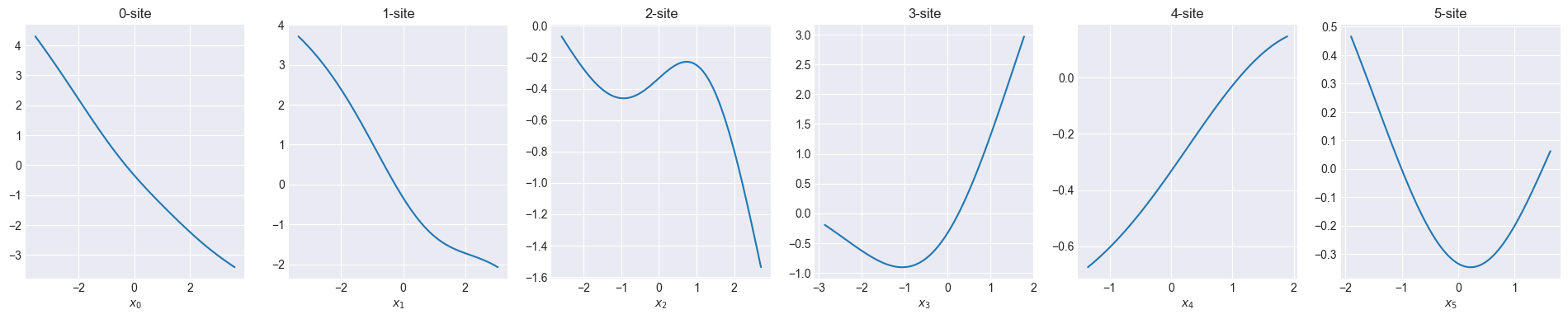
Visualize initial one-body function
Code
def show_onebody(model):
f = model.hidden_size
fig, axs = plt.subplots(1, f, figsize=(4 * f, 4))
for i in range(f):
x = np.linspace(np.min(x_train[:, i]), np.max(x_train[:, i]), 100)
_x = np.zeros((100, 6))
_x[:, i] = x
y = model.forward(_x)[:, 0]
axs[i].set_title(f"{i}-site")
axs[i].set_xlabel(f"$x_{i}$")
axs[i].plot(x, y)
plt.show()Code
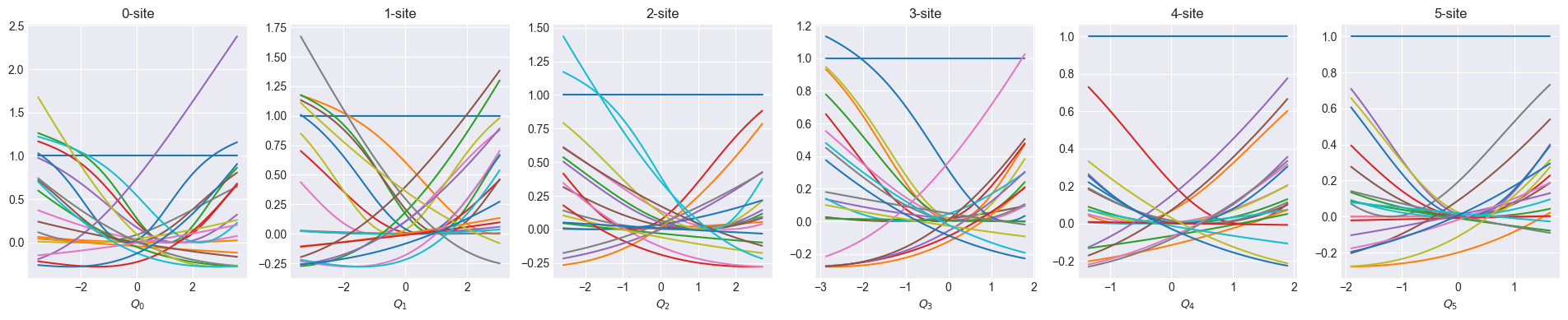
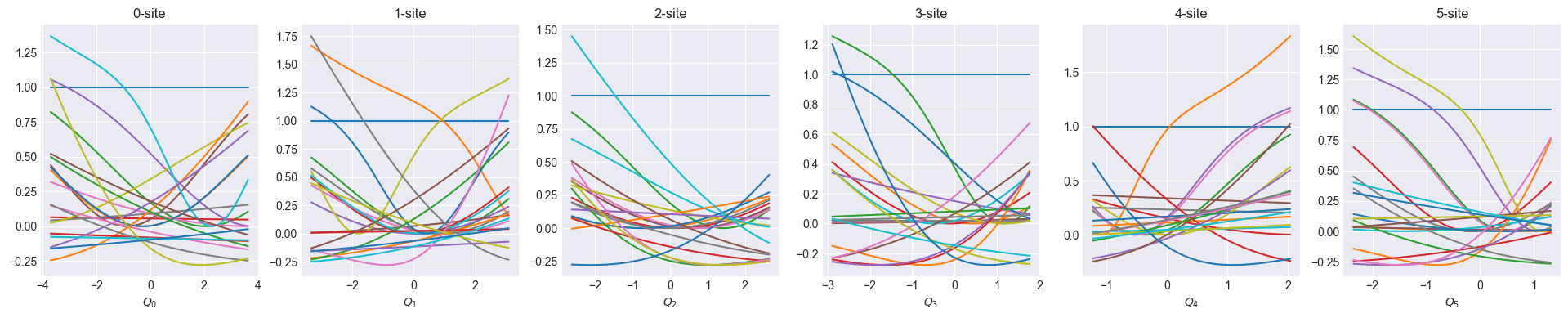
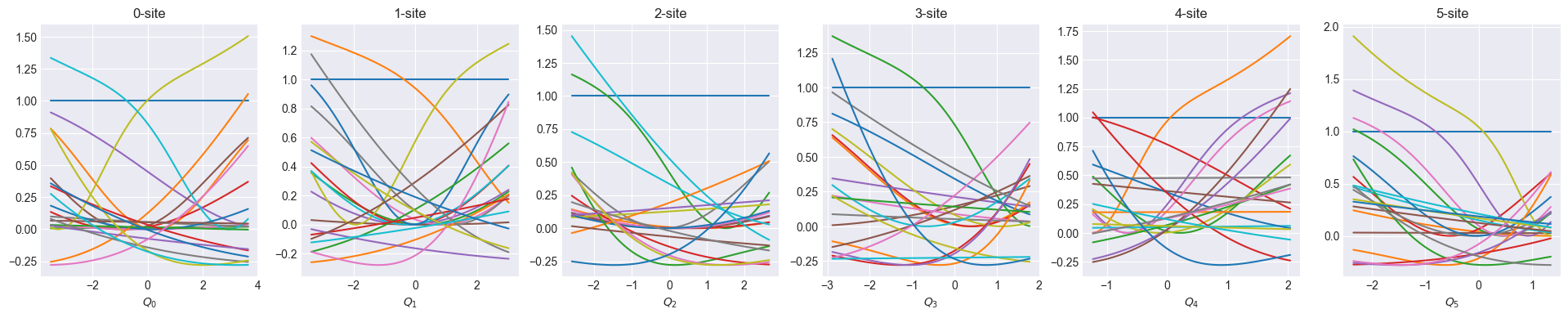
def show_basis(model):
f = model.hidden_size
fig, axs = plt.subplots(1, f, figsize=(4 * f, 4))
q = model.coordinator.forward(x_train)
q0 = model.q0
for i in range(f):
_q = np.linspace(np.min(q[:, i]), np.max(q[:, i]), 100)
phi = model.basis[i].forward(_q, q0[:, i])
axs[i].set_title(f"{i}-site")
axs[i].set_xlabel(f"$Q_{i}$")
axs[i].plot(_q, phi)
plt.show()Optimize one-body
Code
2024-10-16 10:39:44 - INFO:pompon.pompon.model - Model is exported to data/model_initial.h5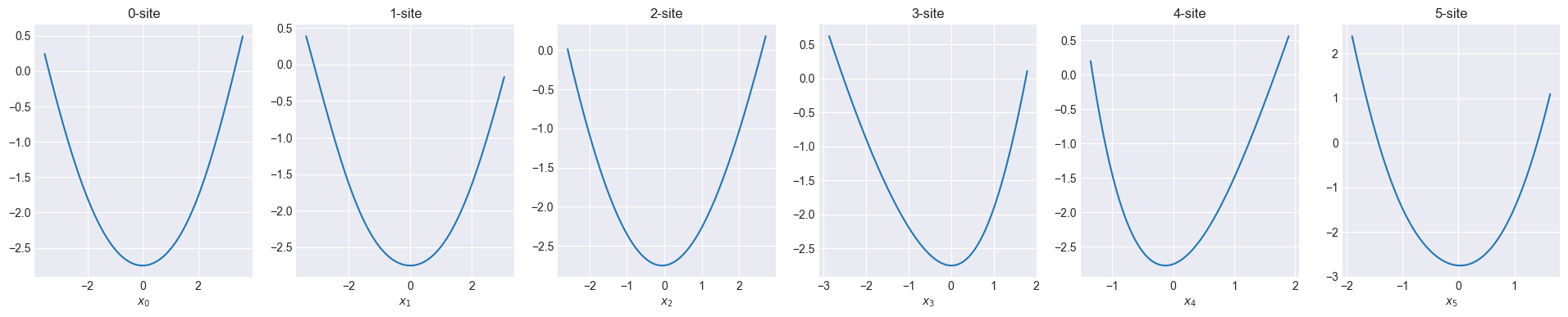
Code
def plot_trace(trace):
plt.plot(
trace["epoch"], np.sqrt(trace["mse_train"]) * y_scale, label="train"
)
plt.plot(trace["epoch"], np.sqrt(trace["mse_test"]) * y_scale, label="test")
plt.xticks(np.arange(0, trace["epoch"][-1], 4000), rotation=45)
plt.xlabel("epochs")
plt.ylabel("RMSE Energy [eV]")
plt.yscale("log")
plt.legend()
plt.show()2024-10-16 10:39:50 - INFO:pompon.pompon.model - Model is exported to data/onebody.h5
2024-10-16 10:39:50 - INFO:pompon.pompon.model - Model is imported from data/onebody.h5Sum of one-body function can fit in 100 meV order.
Visualize coordinator after one-body optimization
Use one-body function as initial NNMPO
\[ y = W_0 + W_1\phi^{(1)} + \cdots W_f\phi^{(f)} \] \[ \to \begin{bmatrix} 1 & W_0 + W_1\phi^{(1)} \\ \end{bmatrix} \begin{bmatrix} 1 & W_2\phi^{(2)} \\ 0 & 1 \\ \end{bmatrix} \begin{bmatrix} 1 & W_3\phi^{(3)} \\ 0 & 1 \\ \end{bmatrix} \cdots \begin{bmatrix} W_3\phi^{(f)} \\ 1 \\ \end{bmatrix} \]
2024-10-16 10:39:51 - INFO:pompon.pompon.sop - OneBody function is converted to NNMPOCode
2024-10-16 10:39:51 - INFO:pompon.pompon.model - Model is exported to data/model_initial.h5Code
import optax
for i in range(5):
optimizer.lr = 1.0e-03
solver = optax.adam(1.0e-04)
sweeper.sweep(
nsweeps=3,
opt_tol=1.0e-10,
opt_maxiter=100,
opt_batchsize=625,
cutoff=1.0e-01,
optax_solver=solver,
onedot=(i == 0), # <- First step should be onedot.
auto_onedot=False, # <- Automatically switch to onedot if the bond dimension reaches maxdim.
maxdim=14,
)
trace = optimizer.optimize(epochs=200, epoch_per_log=200)
nnmpo.export_h5(
f'data/nnmpo_step_{i}_rmse_{np.sqrt(trace["mse_test"][-1]) * y_scale:.3e}.h5'
)
for i in range(5, 10):
optimizer.lr = 1.0e-04 # <- Lower learning rate.
solver = optax.adam(1.0e-05)
sweeper.sweep(
nsweeps=3,
opt_tol=1.0e-10,
opt_maxiter=500,
opt_batchsize=625,
cutoff=1.0e-02,
optax_solver=solver,
onedot=(i == 0), # <- First step should be onedot.
auto_onedot=True, # <- Automatically switch to onedot if the bond dimension reaches maxdim.
maxdim=14,
)
trace = optimizer.optimize(epochs=200, epoch_per_log=200)
nnmpo.export_h5(
f'data/nnmpo_step_{i}_rmse_{np.sqrt(trace["mse_test"][-1]) * y_scale:.3e}.h5'
)
# Conjugate gradient optimization
sweeper.sweep(
nsweeps=10,
opt_tol=1.0e-08,
opt_maxiter=2000,
opt_batchsize=625,
# optax_solver=solver,
use_CG=True, # <-- !!!
onedot=True,
maxdim=14,
)
optimizer.lr = 1.0e-07
trace = optimizer.optimize(epochs=200, epoch_per_log=200)
plot_trace(trace)
show_basis(nnmpo)2024-10-16 10:40:02 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_0_rmse_7.492e-02.h52024-10-16 10:40:16 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_1_rmse_2.839e-02.h52024-10-16 10:40:30 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_2_rmse_2.070e-02.h52024-10-16 10:40:43 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_3_rmse_3.494e-02.h52024-10-16 10:40:56 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_4_rmse_1.418e-02.h52024-10-16 10:41:23 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_5_rmse_7.052e-03.h52024-10-16 10:41:41 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_6_rmse_5.909e-03.h52024-10-16 10:41:59 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_7_rmse_5.411e-03.h52024-10-16 10:42:17 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_8_rmse_5.059e-03.h52024-10-16 10:42:35 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_step_9_rmse_4.715e-03.h5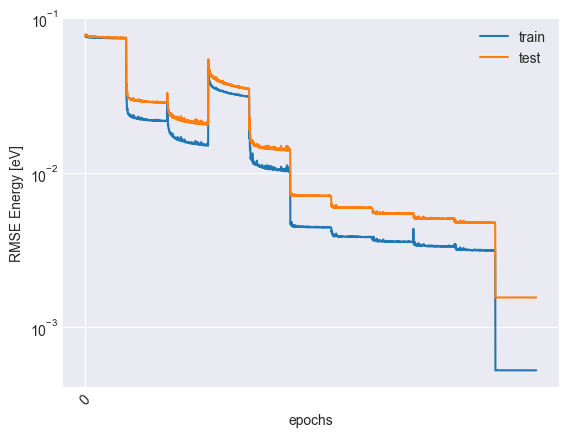
Code
2024-10-16 10:44:16 - INFO:pompon.pompon.model - Model is exported to data/nnmpo_final_rmse_1.550e-03.h5| epoch | mse_train | mse_test | mse_train_f | tt_norm | tt_ranks |
|---|---|---|---|---|---|
| i64 | f64 | f64 | f64 | f64 | list[i64] |
| 1 | 0.033321 | 0.034835 | 0.259389 | -14.813811 | [2, 2, … 2] |
| 2 | 0.033062 | 0.034307 | 0.259242 | -14.813811 | [2, 2, … 2] |
| 3 | 0.033042 | 0.033916 | 0.256926 | -14.813811 | [2, 2, … 2] |
| 4 | 0.032263 | 0.033084 | 0.255528 | -14.813811 | [2, 2, … 2] |
| 5 | 0.033542 | 0.034455 | 0.254564 | -14.813811 | [2, 2, … 2] |
| … | … | … | … | … | … |
| 2196 | 0.000002 | 0.000014 | 0.00001 | -220.626693 | [8, 14, … 9] |
| 2197 | 0.000002 | 0.000013 | 0.00001 | -220.626693 | [8, 14, … 9] |
| 2198 | 0.000002 | 0.000013 | 0.00001 | -220.626693 | [8, 14, … 9] |
| 2199 | 0.000002 | 0.000013 | 0.00001 | -220.626693 | [8, 14, … 9] |
| 2200 | 0.000002 | 0.000013 | 0.00001 | -220.626693 | [8, 14, … 9] |
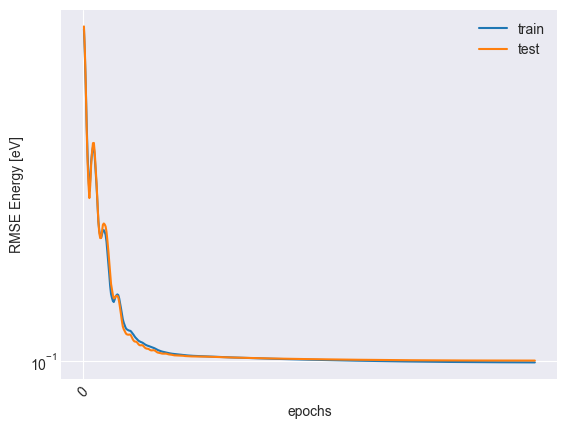
Visualize the optimization traces
Code
plt.plot(trace["epoch"], np.sqrt(trace["mse_train"]) * y_scale, label="train")
plt.plot(trace["epoch"], np.sqrt(trace["mse_test"]) * y_scale, label="test")
plt.xticks(np.arange(0, trace["epoch"][-1], 4000), rotation=45)
plt.xlabel("epochs")
plt.ylabel("RMSE Energy [eV]")
plt.yscale("log")
plt.legend()
plt.savefig('assets/trace.svg')
plt.show()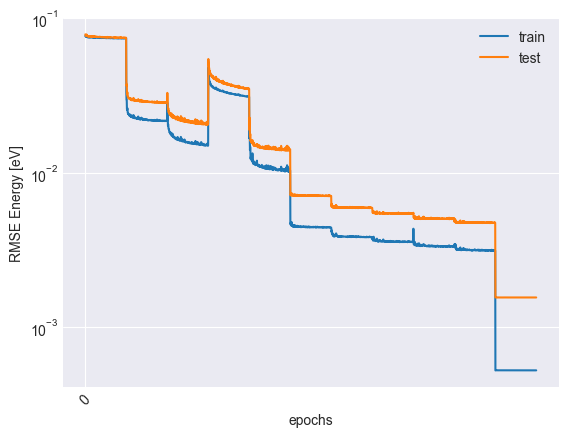
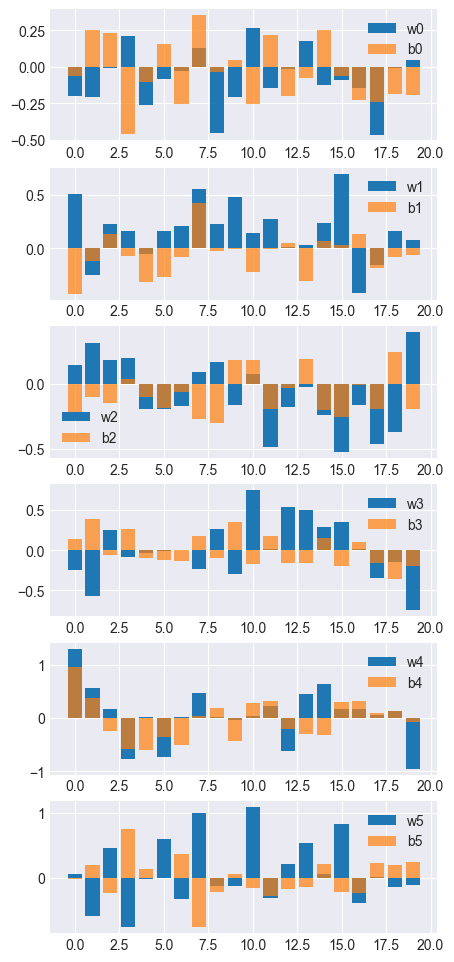
Parameter(w1, shape=(20,))
Parameter(b1, shape=(20,))
[-0.39287908 -0.13464318 0.14050318 -0.07814877 -0.03572304 -0.37963561
-0.0855011 0.55868097 -0.01750081 0.23813855 -0.27342234 0.04030169
0.12940236 -0.32228352 0.09547939 -0.01471205 0.06512873 -0.2840365
0.04007023 0.20726764]
Parameter(b3, shape=(20,))
[ 0.15595645 0.78521664 -0.22730115 0.3919457 -0.26728326 -0.24770266
-0.21508083 0.17693537 -0.05250274 0.41195362 -0.29535957 0.21481374
-0.17795398 -0.26547028 0.2415685 -0.15915255 0.38545226 -0.17031766
-0.66347873 -0.22850493]
Parameter(w3, shape=(20,))
Parameter(w0, shape=(20,))
Parameter(b0, shape=(20,))
[-0.09522305 0.11708057 0.29824885 -0.48003036 -0.09094859 0.09054434
-0.27302538 0.45346682 0.25239392 0.0707163 -0.31713018 0.03327424
-0.12000518 -0.021749 0.10488636 -0.18124672 -0.33535119 -0.46569359
-0.21225473 -0.10930101]
Parameter(b2, shape=(20,))
[-0.32414808 -0.17914898 -0.11512402 0.07283107 -0.11565191 -0.21038414
-0.12420237 -0.38883823 -0.36601631 0.22639764 0.30256851 -0.25292004
0.1423828 0.26743926 -0.22291431 -0.2352676 0.03834339 -0.20269271
0.29183001 -0.21124825]
Parameter(w2, shape=(20,))
Parameter(w5, shape=(20,))
Parameter(b5, shape=(20,))
[-0.08737266 0.1383658 -0.05758187 0.7263479 0.16535198 -0.05600558
0.35295224 -1.14079062 -0.3458684 -0.01694047 -0.28313654 -0.34195386
-0.26088313 -0.194944 0.23606602 -0.30504939 -0.39490075 0.26061878
0.25830287 0.31402083]
Parameter(w4, shape=(20,))
Parameter(b4, shape=(20,))
[ 0.94919174 0.05418159 -0.81424432 -0.72655884 -0.62482164 -0.45372532
-0.76737424 0.09948951 0.21497529 -0.59259597 0.316253 0.30250957
-0.17445118 -0.26831699 -0.28920347 0.30175617 0.30627362 0.10200498
0.17171362 -0.13517611]
Parameter(x0, shape=(20, 6))
Core(shape=(1, 21, 8), leg_names=('β0', 'i1', 'β1'), dtype=float64)
Parameter(norm, shape=())
Core(shape=(1, 21, 8), leg_names=('β0', 'i1', 'β1'), dtype=float64)
Core(shape=(14, 21, 9), leg_names=('β4', 'i5', 'β5'), dtype=float64)
TwodotCore(shape=(1, 21, 21, 14), leg_names=('β0', 'i1', 'i2', 'β2'))
Core(shape=(8, 21, 14), leg_names=('β1', 'i2', 'β2'), dtype=float64)
Core(shape=(14, 21, 14), leg_names=('β2', 'i3', 'β3'), dtype=float64)
Core(shape=(14, 21, 14), leg_names=('β3', 'i4', 'β4'), dtype=float64)
Core(shape=(9, 21, 1), leg_names=('β5', 'i6', 'β6'), dtype=float64)
Parameter(U, shape=(6, 6))Code
2024-10-16 10:44:17 - INFO:pompon.pompon.model - Model is imported from data/nnmpo_final_rmse_1.550e-03.h5Code
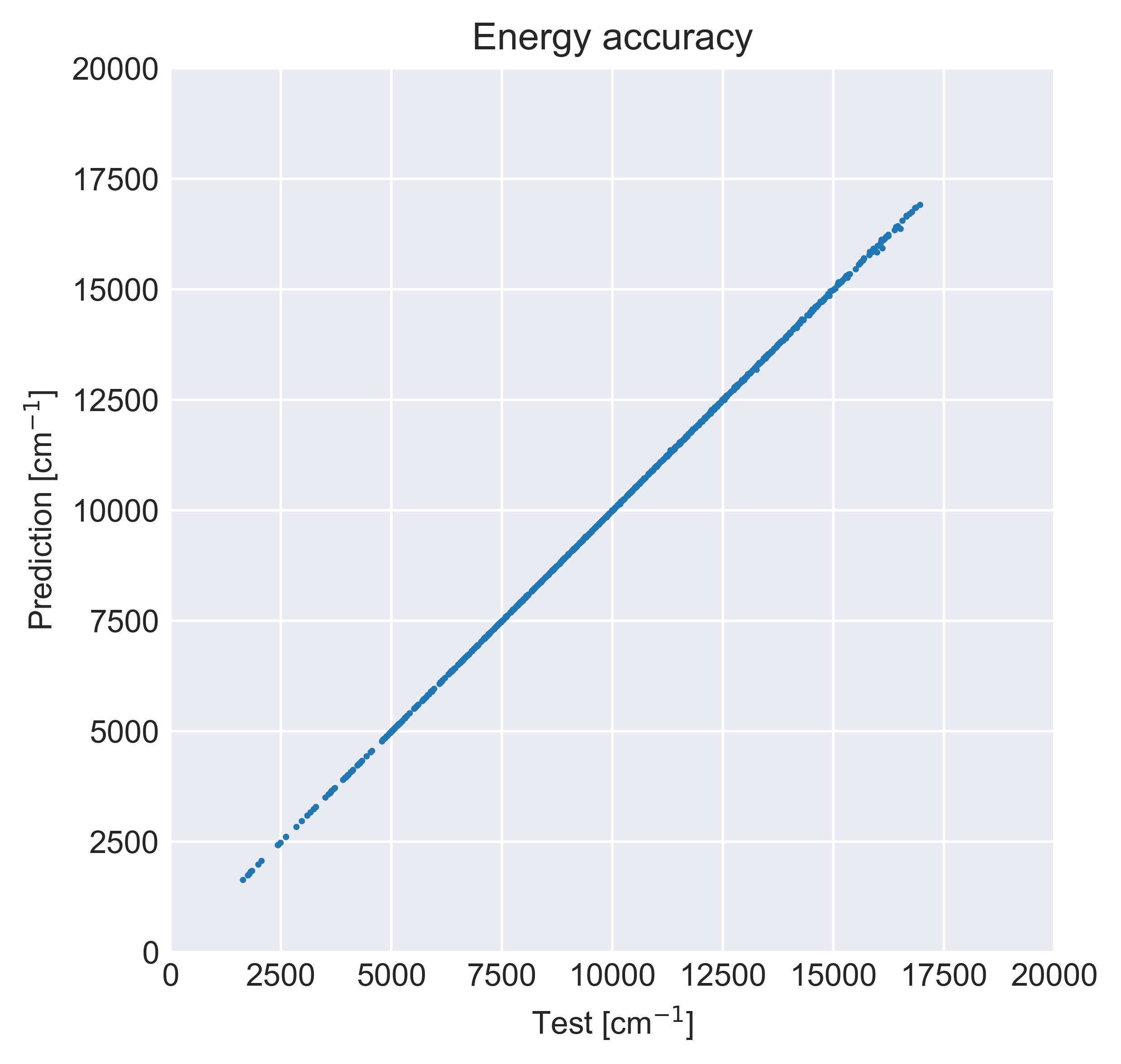
fig, ax = plt.subplots(dpi=500)
ax.scatter(
(y_test * y_scale + y_shift - y_min) * 8.065544e3,
(y_pred + y_shift - y_min) * 8.065544e3,
s=1,
)
plt.gca().set_aspect("equal", adjustable="box")
plt.xlabel("Test [cm$^{-1}$]")
plt.ylabel("Prediction [cm$^{-1}$]")
plt.xlim(0, 20000)
plt.ylim(0, 20000)
plt.title("Energy accuracy")
plt.tight_layout()
plt.show()