3.12.2 (main, Feb 6 2024, 20:19:44) [Clang 15.0.0 (clang-1500.1.0.2.5)]
pompon version = 0.0.9
macOS-14.4.1-arm64-arm-64bitH2CO molecute - visualize
Visualize trained model
Environment
Import modules
Code
import itertools
import matplotlib.pyplot as plt
import numpy as np
import polars as pl
plt.style.use("seaborn-v0_8-darkgrid")
plt.rcParams.update(
{
"image.cmap": "jet",
}
)
x_train = np.load("data/x_train.npy")
y_train = np.load("data/y_train.npy")
x_scale = x_train.std()
y_scale = y_train.std()
y_shift = -3112.1604302407663 # eV (mean)
y_min = -3113.4044750979383 # eV (eq. position)
print(f"{x_scale=}, {y_scale=}")x_scale=np.float64(17.203527572055016), y_scale=np.float64(0.4220226766217427)Load data
coef satisfies \[
Q_i = c_i q_i
\]
where \(Q_i\) is dimensionless, \(q_i\) is mass-weighted with unit \(\sqrt{m_{\mathrm{e}}}\cdot a_0\) and \[ c_i = \sqrt{\frac{\omega_i}{\hbar}} \times {\left[\sqrt{\frac{\mathrm{AMU}}{m_{\mathrm{e}}}}\frac{\mathring{\mathrm{A}}}{a_0}\right]} \]
Visualize 2D cut
Code
df = pl.read_parquet("data/h2co_dft_2MR.parquet")
n = 6
selected_df = df.filter( # (pl.col('Q0') == 0) & (pl.col('Q1') == 0) &
(pl.col("Q2") == 0)
& (pl.col("Q3") == 0)
& (pl.col("Q4") == 0)
& (pl.col("Q5") == 0)
).sort(by=["Q1", "Q0"])
Q0 = np.array(selected_df["Q0"])
Q0 = np.linspace(-3.0, 3.0, 20)
# insert 0.0 in sorted order
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
Q1 = np.array(selected_df["Q1"])
Q1 = np.linspace(-3.0, 3.0, 20)
Q1 = np.insert(Q1, np.searchsorted(Q1, 0.0), 0.0)
min_ener = df.filter(
(pl.col("Q0") == 0)
& (pl.col("Q1") == 0)
& (pl.col("Q2") == 0)
& (pl.col("Q3") == 0)
& (pl.col("Q4") == 0)
& (pl.col("Q5") == 0)
)["ener"][0]
y_ref = np.array(selected_df["ener"]).reshape(21, 21) - min_ener
fig = plt.figure(figsize=(3 * n + 3, 3 * n), dpi=300)
subfigs = [
[fig.add_subplot(n, n + 1, i * (n + 1) + j + 1) for j in range(n)]
for i in range(n)
]
vmax = 2.0
levels = np.arange(-0.1, 2.05, 0.05)
axs = subfigs[0][1]
im = axs.contourf(
Q0,
Q1,
y_ref,
50,
vmax=vmax,
vmin=0.0,
levels=levels,
)
cbar_ax = fig.add_axes([0.85, 0.15, 0.01, 0.7])
fig.colorbar(im, cax=cbar_ax, label="Energy [eV]")
active_modes = set(range(n))
for dims in itertools.combinations(range(n), 2):
fixed_modes = list(active_modes - set(dims))
target_modes = list(dims)
Q0 = np.linspace(-3.0, 3.0, 20)
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
Q1 = np.linspace(-3.0, 3.0, 20)
Q1 = np.insert(Q1, np.searchsorted(Q1, 0.0), 0.0)
Q0, Q1 = np.meshgrid(Q0, Q1)
x = np.zeros((21 * 21, n))
x[:, dims[0]] = Q0.flatten() / coef[dims[0]]
x[:, dims[1]] = Q1.flatten() / coef[dims[1]]
y = (
nnmpo.forward(x).reshape(Q0.shape) + y_shift - y_min
) # * y_scale# * 27.21138 # Eh to eV
title = f"cut = {dims} pred"
axs = subfigs[dims[0]][dims[1]]
axs.set_yticks([-2, 0, 2])
axs.set_xticks([-2, 0, 2])
axs.set_title(title)
im = axs.contourf(
Q0,
Q1,
y,
50,
vmax=vmax,
vmin=0,
levels=levels,
)
for dims in itertools.combinations(range(n), 2):
fixed_modes = list(active_modes - set(dims))
target_modes = list(dims)
selected_df = df.filter(
(pl.col(f"Q{fixed_modes[0]}") == 0)
& (pl.col(f"Q{fixed_modes[1]}") == 0)
& (pl.col(f"Q{fixed_modes[2]}") == 0)
& (pl.col(f"Q{fixed_modes[3]}") == 0)
).sort(by=[f"Q{target_modes[1]}", f"Q{target_modes[0]}"])
Q0 = np.array(selected_df[f"Q{target_modes[0]}"])
Q0 = np.linspace(-3.0, 3.0, 20)
# insert 0.0 in sorted order
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
Q1 = np.array(selected_df[f"Q{target_modes[1]}"])
Q1 = np.linspace(-3.0, 3.0, 20)
Q1 = np.insert(Q1, np.searchsorted(Q1, 0.0), 0.0)
y = np.array(selected_df["ener"]).reshape(21, 21) - min_ener
title = f"cut = {dims} true"
axs = subfigs[dims[1]][dims[0]]
axs.set_yticks([-2, 0, 2])
axs.set_xticks([-2, 0, 2])
axs.set_title(title)
im = axs.contourf(
Q0,
Q1,
y,
50,
vmax=vmax,
vmin=0,
levels=levels,
)
for dim in range(n):
axs = subfigs[dim][dim]
axs.clear()
x = np.zeros((21, n))
Q0 = np.linspace(-3.0, 3.0, 20)
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
x[:, dim] = Q0 / coef[dim]
y = (
nnmpo.forward(x).reshape(-1) + y_shift - y_min
) # * y_scale # * 27.21138 # Eh to eV
label = "pred" if dim == 0 else None
axs.plot(Q0, y, label=label)
Q0 = np.linspace(-3.0, 3.0, 20)
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
fixed_modes = list(active_modes - set([dim]))
selected_df = df.filter(
(pl.col(f"Q{fixed_modes[0]}") == 0)
& (pl.col(f"Q{fixed_modes[1]}") == 0)
& (pl.col(f"Q{fixed_modes[2]}") == 0)
& (pl.col(f"Q{fixed_modes[3]}") == 0)
& (pl.col(f"Q{fixed_modes[4]}") == 0)
).sort(by=[f"Q{dim}"])
y = np.array(selected_df["ener"]) - min_ener
label = "true" if dim == 0 else None
axs.plot(Q0, y, label=label)
if dim == 0:
axs.legend()
plt.savefig("data/h2co_2dpes.png")
plt.show()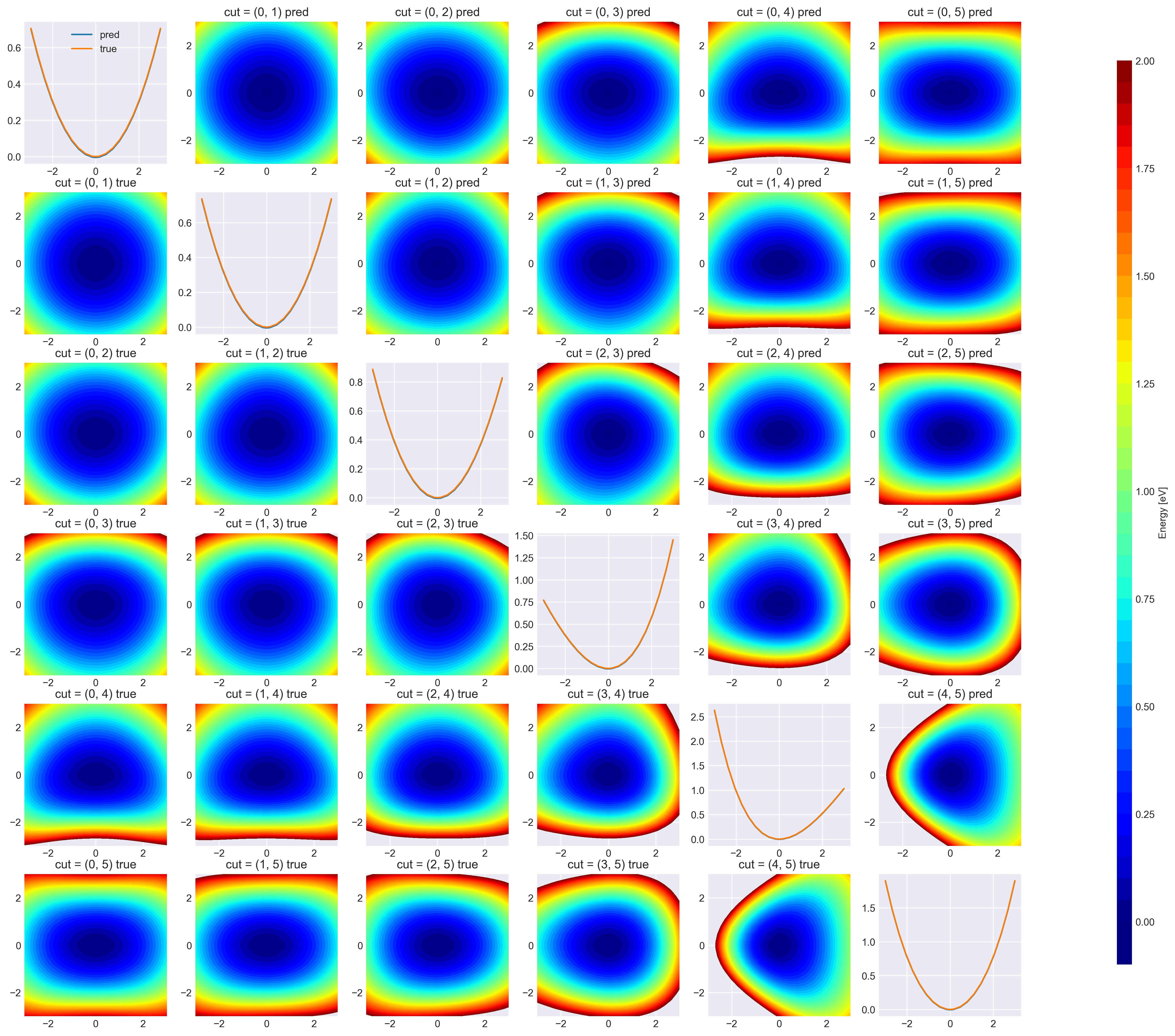
Visualize coordinator
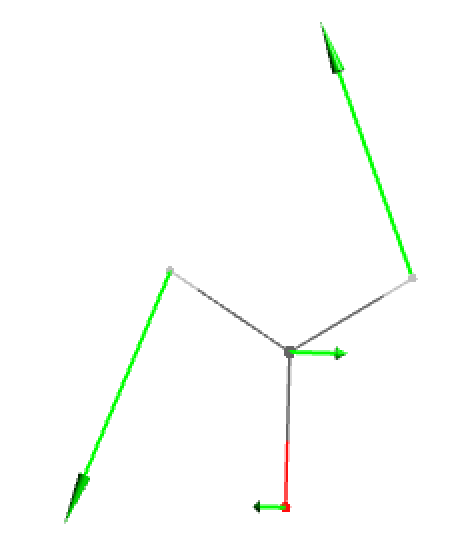
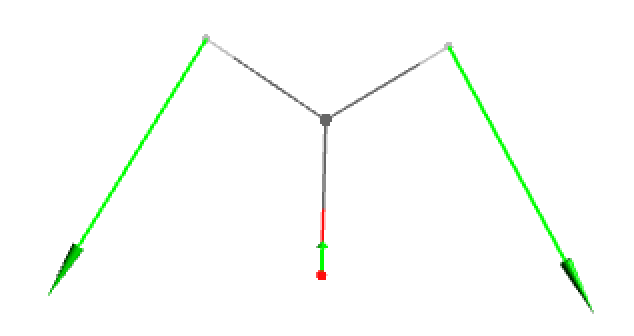
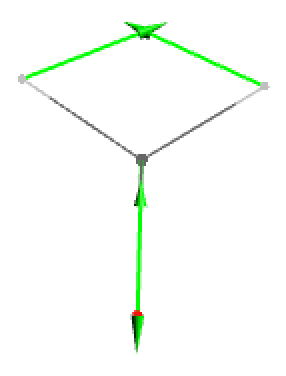
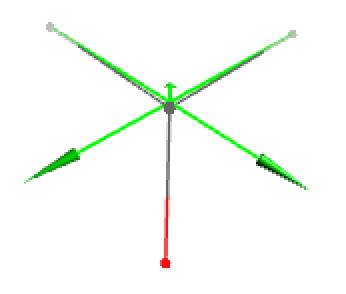
- Original displacement vectors
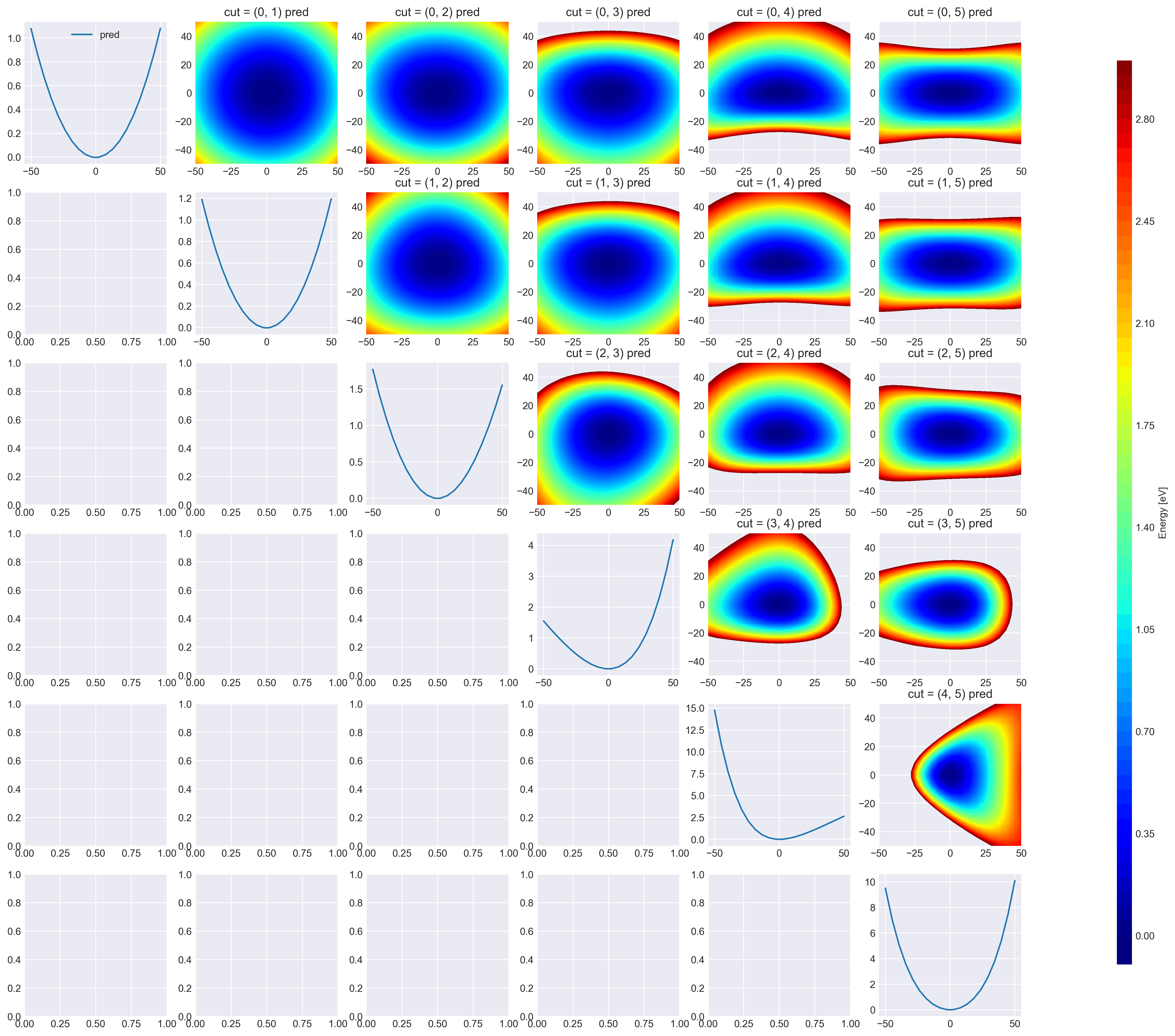
- Visualize 2D cut of latent DOFs
Code
n = 6
fig = plt.figure(figsize=(3 * n + 3, 3 * n), dpi=300)
subfigs = [
[fig.add_subplot(n, n + 1, i * (n + 1) + j + 1) for j in range(n)]
for i in range(n)
]
vmax = 3.0
# cmap = "plasma" #'viridis' # 'RdGy'
levels = np.arange(-0.1, 3.05, 0.05)
axs = subfigs[1][0] # .subplots()
im = axs.contourf(
Q0,
Q1,
y_ref,
50,
vmax=vmax,
vmin=0.0,
levels=levels, # cmap=cmap,
)
cbar_ax = fig.add_axes([0.85, 0.15, 0.01, 0.7])
fig.colorbar(im, cax=cbar_ax, label="Energy [eV]")
active_modes = set(range(n))
for dims in itertools.combinations(range(n), 2):
fixed_modes = list(active_modes - set(dims))
target_modes = list(dims)
Q0 = np.linspace(-50.0, 50.0, 20)
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
Q1 = np.linspace(-50.0, 50.0, 20)
Q1 = np.insert(Q1, np.searchsorted(Q1, 0.0), 0.0)
Q0, Q1 = np.meshgrid(Q0, Q1)
x = np.zeros((21 * 21, n))
x += 0.0 # cut of 10.0
x[:, dims[0]] = Q0.flatten()
x[:, dims[1]] = Q1.flatten()
# x /= x_scale
y = (
nnmpo.tt.forward(nnmpo.basis.forward(x, nnmpo.q0)).reshape(Q0.shape)
+ y_shift
- y_min
) # * y_scale
title = f"cut = {dims} pred"
axs = subfigs[dims[0]][dims[1]] # .subplots()
# axs.set_yticks([-2, 0, 2])
# axs.set_xticks([-2, 0, 2])
axs.set_title(title)
im = axs.contourf(
Q0,
Q1,
y,
50,
vmax=vmax,
vmin=0,
levels=levels, # , cmap=cmap
)
for dim in range(n):
axs = subfigs[dim][dim]
axs.clear()
x = np.zeros((21, n))
Q0 = np.linspace(-50.0, 50.0, 20)
Q0 = np.insert(Q0, np.searchsorted(Q0, 0.0), 0.0)
x[:, dim] = Q0 # / x_scale
y = (
nnmpo.tt.forward(nnmpo.basis.forward(x, nnmpo.q0)).reshape(-1)
+ y_shift
- y_min
) # * y_scale
label = "pred" if dim == 0 else None
axs.plot(Q0, y, label=label)
if dim == 0:
axs.legend()
subfigs[1][0].clear()
plt.savefig("data/h2co_2dpes_latent.png")
plt.show()