Code
3.12.2 (main, Feb 6 2024, 20:19:44) [Clang 15.0.0 (clang-1500.1.0.2.5)]
pompon version = 0.0.9
macOS-14.4.1-arm64-arm-64bitIn this example we will consider \[ V(\boldsymbol{x}) = \boldsymbol{x}^\intercal \boldsymbol{A} \boldsymbol{x} \] where \(\boldsymbol{A}\) satisfies \(\det{\boldsymbol{A}}>0\).
A=
[[3. 1. 1. 1. 1. 1. ]
[1. 3.6 1. 1. 1. 1. ]
[1. 1. 4.2 1. 1. 1. ]
[1. 1. 1. 4.8 1. 1. ]
[1. 1. 1. 1. 5.4 1. ]
[1. 1. 1. 1. 1. 6. ]]def plot_2d_cut(dims=(0, 1)):
# cut of z
q0 = np.linspace(-1, 1, 100)
q1 = np.linspace(-1, 1, 100)
Q0, Q1 = np.meshgrid(q0, q1)
x = np.zeros((100 * 100, n))
x[:, dims[0]] = Q0.flatten()
x[:, dims[1]] = Q1.flatten()
y = ellipse_pes(x).reshape(Q0.shape)
plt.clf()
plt.contourf(Q0, Q1, y, 50, cmap="RdGy", vmax=9, vmin=0)
plt.xlabel(f"Q{dims[0]}")
plt.ylabel(f"Q{dims[1]}")
plt.title("2D-cut")
plt.colorbar()
plt.tight_layout()
plt.show()Learning points are sampled from Boltzman distribution \(\exp(-\beta V)\), which is anallytically achievable by using multivariate normal distribution.
dims = (0, 1)
q0 = np.linspace(-1, 1, 100)
q1 = np.linspace(-1, 1, 100)
Q0, Q1 = np.meshgrid(q0, q1)
x = np.zeros((100 * 100, n))
x[:, dims[0]] = Q0.flatten()
x[:, dims[1]] = Q1.flatten()
y = ellipse_pes(x).reshape(Q0.shape)
plt.contourf(Q0, Q1, y, 50, cmap="RdGy", vmax=9, vmin=0)
plt.colorbar()
plt.xlabel(f"Q{dims[0]}")
plt.ylabel(f"Q{dims[1]}")
x = sample_ellipse(N=100, β=1.0e01)
plt.scatter(x[:, dims[0]], x[:, dims[1]], label="β=1.e+01")
x = sample_ellipse(N=100, β=1.0e00)
plt.scatter(x[:, dims[0]], x[:, dims[1]], label="β=1.e+00")
plt.legend()
plt.show()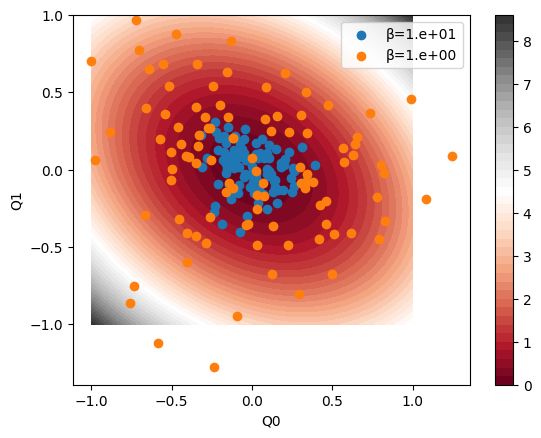
One will sample \(20n^2\) points with temperature \(\beta=1.0\).
(503, 6)
(503, 1)
(503, 6)To stabilize the learning, one standardizes the input data.
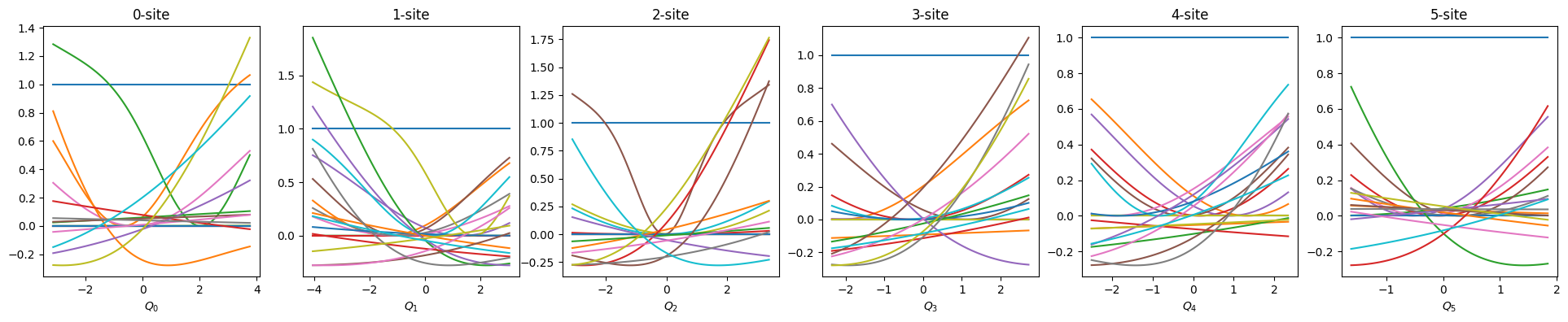
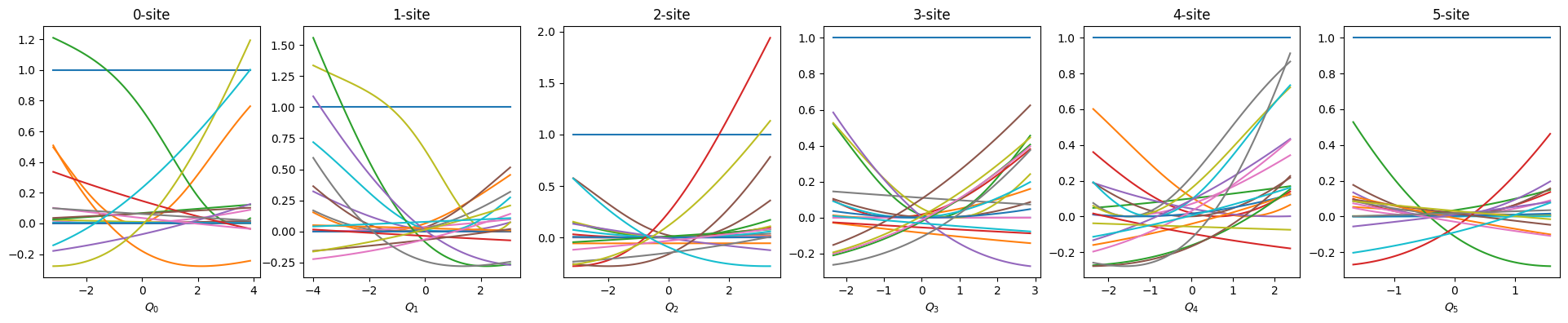
def show_basis(model):
f = model.hidden_size
fig, axs = plt.subplots(1, f, figsize=(4 * f, 4))
q = model.coordinator.forward(x_train)
q0 = model.q0
for i in range(f):
_q = np.linspace(np.min(q[:, i]), np.max(q[:, i]), 100)
phi = model.basis[i].forward(_q, q0[:, i])
axs[i].set_title(f"{i}-site")
axs[i].set_xlabel(f"$Q_{i}$")
axs[i].plot(_q, phi)
plt.show()
show_basis(nnmpo)
nnmpo.basis.plot_data()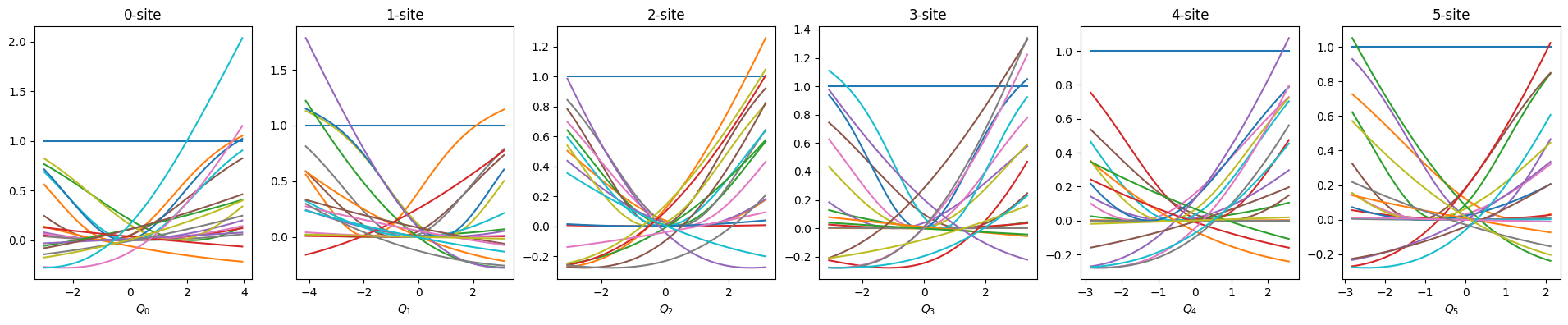
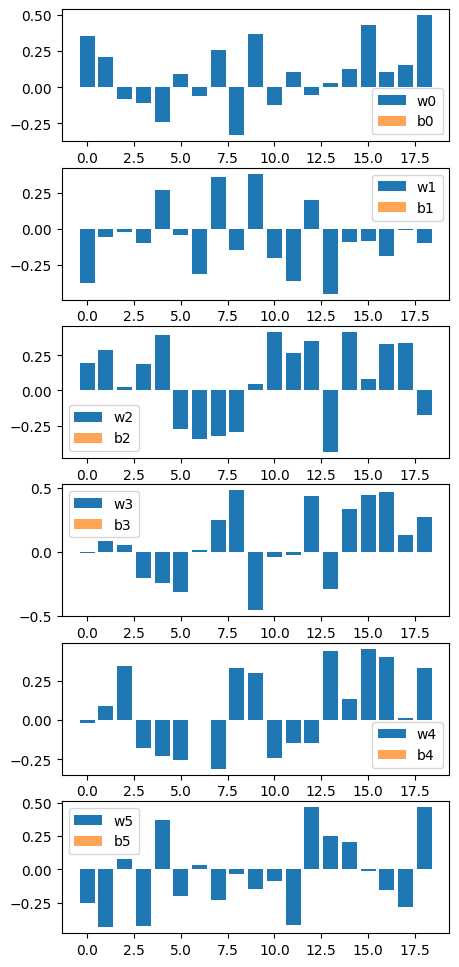
Construct model like this;
Let’s start machine learning!
Adam: Adam optimizerlr: learning rate \(\eta\)batch_size: Batch size for stochastic gradient descent.epochs: Number of iterations for optimizing \(w\), \(b\), and \(U\).nsweeps: Number of sweepsoptax_solver: Solver used for TT optimizationcutoff: Singular value cutoff theresholdmaxdim: Max bond dimensionWe know that \[ V(\boldsymbol{x}) = \boldsymbol{x}^\intercal \boldsymbol{A} \boldsymbol{x} \] has 2-rank structure by diagonalizing \(\boldsymbol{A}\), i.e., \[ V(\boldsymbol{x}) = \boldsymbol{x}^\intercal \boldsymbol{U} \boldsymbol{\Lambda} \boldsymbol{U}^\intercal \boldsymbol{x} = \boldsymbol{q}^\intercal \boldsymbol{\Lambda} \boldsymbol{q} = \begin{bmatrix} 1 & \lambda_1 q_1^2 \end{bmatrix} \begin{bmatrix} 1 & \lambda_2 q_2^2 \\ 0 & 1 \end{bmatrix} \cdots \begin{bmatrix} 1 & \lambda_{n-1} q_{n-1}^2 \\ 0 & 1 \end{bmatrix} \begin{bmatrix} \lambda_n q_n^2 \\ 1 \end{bmatrix} \]
Thus, we set maxdim=2.
import optax
from pompon.optimizer import Adam, Sweeper
optimizer = Adam(lr=1.0e-02).setup(
model=nnmpo,
x_train=x_train,
y_train=y_train,
f_train=f_train,
batch_size=125,
x_test=x_test,
y_test=y_test,
)
sweeper = Sweeper(optimizer)
for i in range(4):
trace = optimizer.optimize(
epochs=200,
epoch_per_log=200,
)
show_basis(nnmpo)
trace = sweeper.sweep(
nsweeps=4,
optax_solver=optax.adam(1.0e-03),
opt_batchsize=10000,
opt_maxiter=1000,
opt_tol=1.0e-03,
cutoff=1.0e-01,
maxdim=2,
onedot=(i == 0),
auto_onedot=True,
)
# After adam, switch to conjugate gradient.
trace = sweeper.sweep(
nsweeps=10,
use_CG=True,
opt_batchsize=10000,
opt_maxiter=1000,
opt_tol=1.0e-10,
onedot=True,
)
optimizer.lr = 1.0e-05 # reduce learning rate
trace = optimizer.optimize(
epochs=200,
epoch_per_log=200,
)2024-10-16 10:37:11 - INFO:pompon.pompon.model - Model is exported to ./model_initial.h5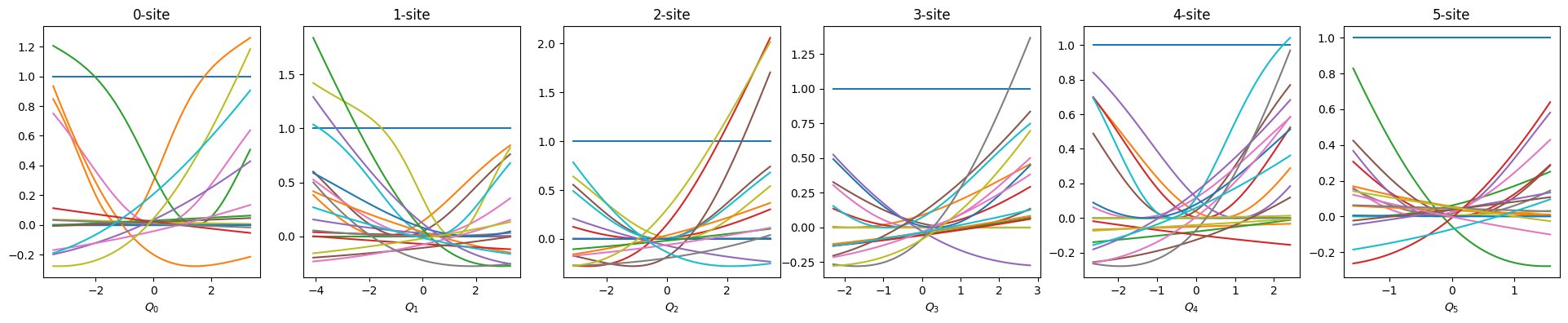

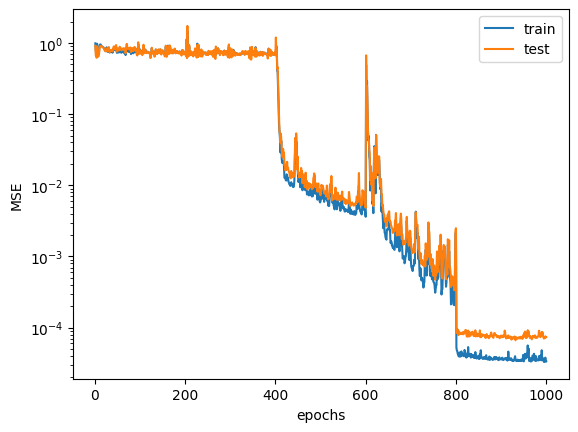
| epoch | mse_train | mse_test | mse_train_f | tt_norm | tt_ranks |
|---|---|---|---|---|---|
| i64 | f64 | f64 | f64 | f64 | list[i64] |
| 1 | 0.989216 | 0.913552 | 2.601824 | 1.0 | [1, 1, … 1] |
| 2 | 0.787074 | 0.670019 | 2.085504 | 1.0 | [1, 1, … 1] |
| 3 | 0.719287 | 0.653636 | 1.756305 | 1.0 | [1, 1, … 1] |
| 4 | 0.711022 | 0.620294 | 1.647353 | 1.0 | [1, 1, … 1] |
| 5 | 0.975683 | 0.895574 | 1.411286 | 1.0 | [1, 1, … 1] |
| … | … | … | … | … | … |
| 996 | 0.000035 | 0.000074 | 0.000101 | -8.3396e7 | [2, 2, … 2] |
| 997 | 0.000033 | 0.000072 | 0.000099 | -8.3396e7 | [2, 2, … 2] |
| 998 | 0.000038 | 0.000076 | 0.000102 | -8.3396e7 | [2, 2, … 2] |
| 999 | 0.000034 | 0.000073 | 0.0001 | -8.3396e7 | [2, 2, … 2] |
| 1000 | 0.000034 | 0.000074 | 0.000101 | -8.3396e7 | [2, 2, … 2] |
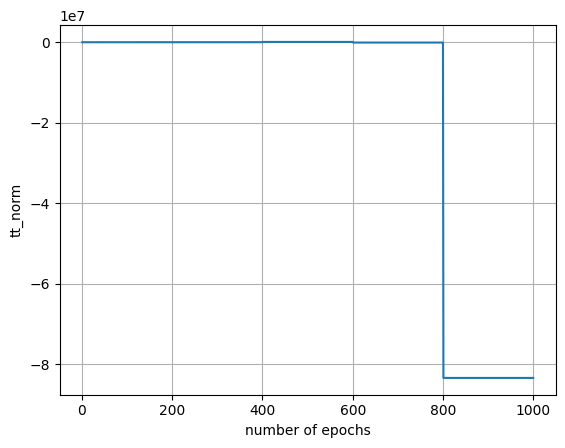
| epoch | mse_train | mse_test | mse_train_f | tt_norm | tt_ranks |
|---|---|---|---|---|---|
| i64 | f64 | f64 | f64 | f64 | list[i64] |
| 996 | 0.000035 | 0.000074 | 0.000101 | -8.3396e7 | [2, 2, … 2] |
| 997 | 0.000033 | 0.000072 | 0.000099 | -8.3396e7 | [2, 2, … 2] |
| 998 | 0.000038 | 0.000076 | 0.000102 | -8.3396e7 | [2, 2, … 2] |
| 999 | 0.000034 | 0.000073 | 0.0001 | -8.3396e7 | [2, 2, … 2] |
| 1000 | 0.000034 | 0.000074 | 0.000101 | -8.3396e7 | [2, 2, … 2] |
Before visualizing, we rescale model parameters to be consistent with the original scale because we trained with standardized data.
import itertools
q0 = np.linspace(-1, 1, 100)
q1 = np.linspace(-1, 1, 100)
Q0, Q1 = np.meshgrid(q0, q1)
fig, ax = plt.subplots(n, n, figsize=(3 * n, 3 * n), dpi=300)
x = np.zeros((100 * 100, n))
x[:, 0] = Q0.flatten()
x[:, 1] = Q1.flatten()
y = ellipse_pes(x).reshape(Q0.shape)
im = ax[0, 0].contourf(Q0, Q1, y, 50, cmap="RdGy", vmax=9, vmin=0)
cbar_ax = fig.add_axes([0.92, 0.15, 0.02, 0.7])
fig.colorbar(im, cax=cbar_ax)
for dims in itertools.combinations(range(n), 2):
x = np.zeros((100 * 100, n))
x[:, dims[0]] = Q0.flatten()
x[:, dims[1]] = Q1.flatten()
y = nnmpo.forward(x).reshape(Q0.shape) + y_shift
title = f"cut = {dims} pred"
ax[dims[0], dims[1]].set_title(title)
im = ax[dims[0], dims[1]].contourf(
Q0, Q1, y, 50, cmap="RdGy", vmax=9, vmin=0
)
for dims in itertools.combinations(range(n), 2):
x = np.zeros((100 * 100, n))
x[:, dims[0]] = Q0.flatten()
x[:, dims[1]] = Q1.flatten()
y = ellipse_pes(x).reshape(Q0.shape)
title = f"cut = {dims} true"
ax[dims[1], dims[0]].set_title(title)
im = ax[dims[1], dims[0]].contourf(
Q0, Q1, y, 50, cmap="RdGy", vmax=9, vmin=0
)
for dim in range(n):
ax[dim, dim].clear()
x = np.zeros((100 * 1, n))
x[:, dim] = q0
y = nnmpo.forward(x).reshape(-1) + y_shift
label = "pred" if dim == 0 else None
ax[dim, dim].plot(q0, y, label=label)
y = ellipse_pes(x).reshape(-1)
label = "true" if dim == 0 else None
ax[dim, dim].plot(q0, y, label=label)
if dim == 0:
ax[dim, dim].legend()
title = (
f"Fitted with N={N}, M={max(trace['tt_ranks'][-1])}, "
+ f"loss={trace['mse_test'][-1]:.3e}"
)
fig.suptitle(title)
plt.savefig("data/6d_pes.png")
plt.show()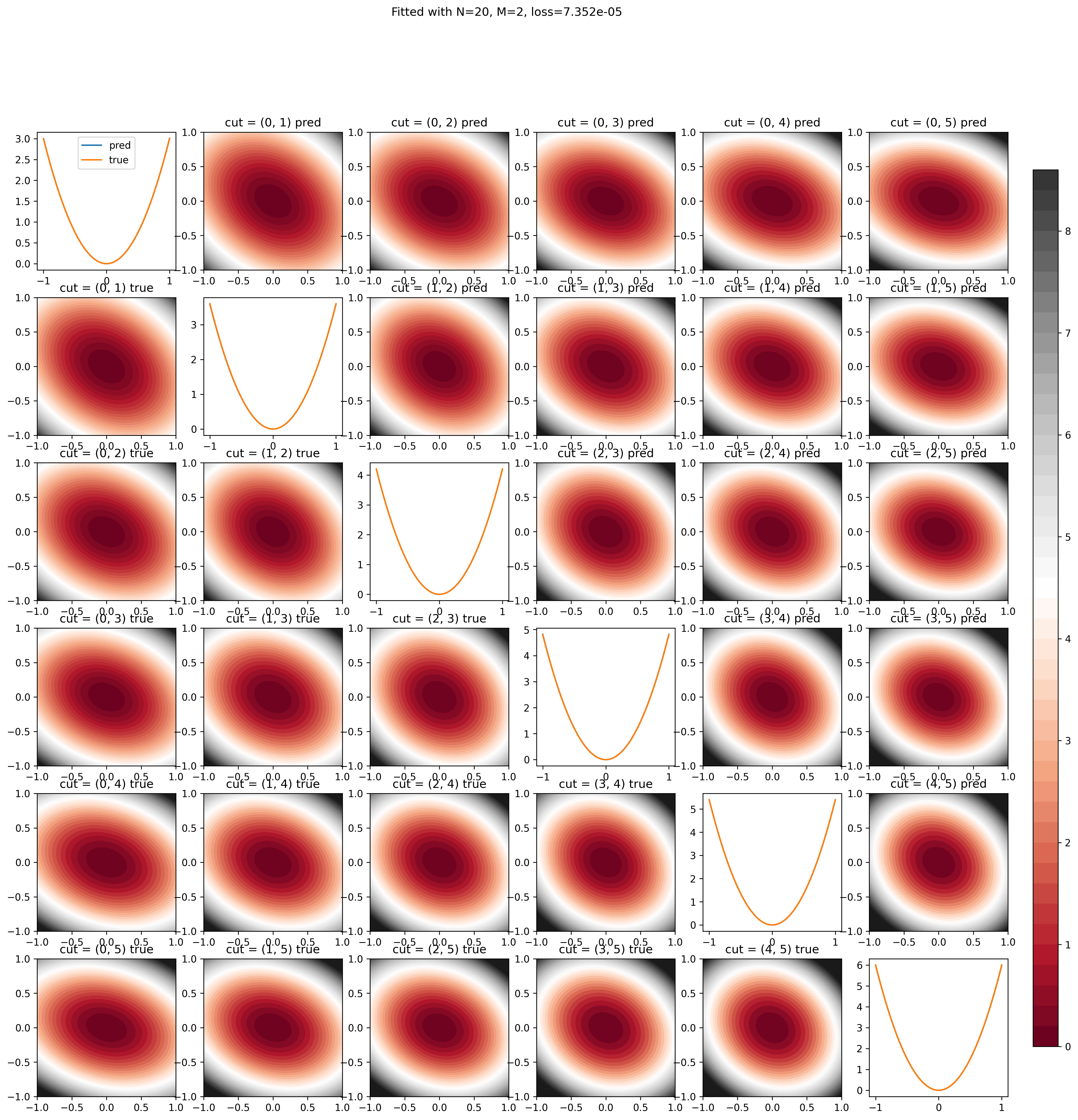
Visualize learning paramter. (chech over fitting occurs or not)